-Search query
-Search result
Showing 1 - 50 of 142 items for (author: namba & k)

EMDB-36190: 
Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-36191: 
Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8jej: 
Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8jek: 
Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-34871: 
Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2
Method: single particle / : Akiba H, Fujita J, Ise T, Nishiyama K, Miyata T, Kato T, Namba K, Ohno H, Kamada H, Nagata S, Tsumoto K

PDB-8hlb: 
Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2
Method: single particle / : Akiba H, Fujita J, Ise T, Nishiyama K, Miyata T, Kato T, Namba K, Ohno H, Kamada H, Nagata S, Tsumoto K
EMDB-33972: 
Cryo-EM structure of the SARS-CoV-2 spike protein (3-up RBD) bound to neutralizing antibody CSW1-1805
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
EMDB-33973: 
Cryo-EM structure of the SARS-CoV-2 spike protein (1-up RBD) bound to neutralizing antibody CSW2-1353
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
EMDB-33974: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing antibody CSW2-1353
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y
EMDB-33975: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) with C480A mutation
Method: single particle / : Anzai I, Fujita J, Ono C, Kosaka Y, Miyamoto Y, Shichinohe S, Takada K, Torii S, Taguwa S, Suzuki K, Makino F, Kajita T, Inoue T, Namba K, Watanabe T, Matsuura Y

EMDB-34429: 
Cryo-EM structure of KpFtsZ-monobody double helical tube
Method: helical / : Fujita J, Amesaka H, Yoshizawa T, Kuroda N, Kamimura N, Hara M, Inoue T, Namba K, Tanaka S, Matsumura H

PDB-8h1o: 
Cryo-EM structure of KpFtsZ-monobody double helical tube
Method: helical / : Fujita J, Amesaka H, Yoshizawa T, Kuroda N, Kamimura N, Hara M, Inoue T, Namba K, Tanaka S, Matsumura H

EMDB-34368: 
Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-34369: 
Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8gy2: 
Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-8gy3: 
Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans
Method: single particle / : Adachi T, Miyata T, Makino F, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-35344: 
Cryo-EM structure of KpFtsZ single filament
Method: helical / : Fujita J, Amesaka H, Yoshizawa T, Kuroda N, Kamimura N, Hibino K, Konishi T, Kato Y, Hara M, Inoue T, Namba K, Tanaka S, Matsumura H

PDB-8ibn: 
Cryo-EM structure of KpFtsZ single filament
Method: helical / : Fujita J, Amesaka H, Yoshizawa T, Kuroda N, Kamimura N, Hibino K, Konishi T, Kato Y, Hara M, Inoue T, Namba K, Tanaka S, Matsumura H

EMDB-34106: 
GroEL on EG-grid stored for 3 months after graphene oxidation
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T
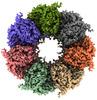
EMDB-34743: 
GroEL on Quantifoil grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T

EMDB-32764: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F

PDB-7wsq: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F, Miyata T, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-32765: 
Cryo-EM structure of the barley Yellow stripe 1 transporter
Method: single particle / : Yamagata A

EMDB-32766: 
Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-DMA
Method: single particle / : Yamagata A

EMDB-32767: 
Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-PDMA
Method: single particle / : Yamagata A

PDB-7wsr: 
Cryo-EM structure of the barley Yellow stripe 1 transporter
Method: single particle / : Yamagata A

PDB-7wst: 
Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-DMA
Method: single particle / : Yamagata A

PDB-7wsu: 
Cryo-EM structure of the barley Yellow stripe 1 transporter in complex with Fe(III)-PDMA
Method: single particle / : Yamagata A

EMDB-34031: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing antibody NIBIC-71
Method: single particle / : Otsubo R, Minamitani T, Kobiyama K, Fujita J, Ito T, Ueno S, Anzai I, Tanino H, Aoyama H, Matsuura Y, Namba K, Imadome K, Ishii KJ, Tsumoto K, Kamitani W, Yasui T

EMDB-34032: 
Cryo-EM structure of the SARS-CoV-2 spike protein (1-up RBD) bound to neutralizing antibody 7G7.2
Method: single particle / : Otsubo R, Minamitani T, Kobiyama K, Fujita J, Ito T, Ueno S, Anzai I, Tanino H, Aoyama H, Matsuura Y, Namba K, Imadome K, Ishii KJ, Tsumoto K, Kamitani W, Yasui T

EMDB-32262: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F, Miyata T, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-7w2j: 
Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus
Method: single particle / : Suzuki Y, Makino F, Miyata T, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-32159: 
GroEL on hydrophilized graphene grid (high particle density)
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T
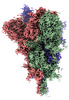
EMDB-32160: 
SARS-CoV-2 spike protein (1-up RBD) on EG-grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T
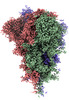
EMDB-32161: 
SARS-CoV-2 spike protein (1-up RBD) on Quantifoil grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T
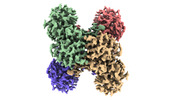
EMDB-32162: 
GAPDH on EG-grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T

EMDB-31605: 
Cryo-EM structure of cyanobacterial photosystem I in the presence of ferredoxin and cytochrome c6
Method: single particle / : Li J, Kurisu G

PDB-7fix: 
Cryo-EM structure of cyanobacterial photosystem I in the presence of ferredoxin and cytochrome c6
Method: single particle / : Li J, Kurisu G
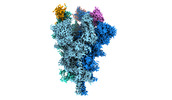
EMDB-32078: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86
Method: single particle / : Maeda R, Fujita J, Konishi Y, Kazuma Y, Yamazaki H, Anzai I, Yamaguchi K, Kasai K, Nagata K, Yamaoka Y, Miyakawa K, Ryo A, Shirakawa K, Makino F, Matsuura Y, Inoue T, Imura A, Namba K, Takaori-Kondo A
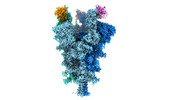
EMDB-32079: 
Cryo-EM structure of the SARS-CoV-2 spike protein (3-up RBD) bound to neutralizing nanobodies P86
Method: single particle / : Maeda R, Fujita J, Konishi Y, Kazuma Y, Yamazaki H, Anzai I, Yamaguchi K, Kasai K, Nagata K, Yamaoka Y, Miyakawa K, Ryo A, Shirakawa K, Makino F, Matsuura Y, Inoue T, Imura A, Namba K, Takaori-Kondo A
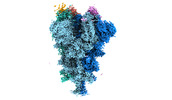
EMDB-32080: 
Cryo-EM structure of the SARS-CoV-2 spike protein (1-up RBD) bound to neutralizing nanobodies P17
Method: single particle / : Maeda R, Fujita J, Konishi Y, Kazuma Y, Yamazaki H, Anzai I, Yamaguchi K, Kasai K, Nagata K, Yamaoka Y, Miyakawa K, Ryo A, Shirakawa K, Makino F, Matsuura Y, Inoue T, Imura A, Namba K, Takaori-Kondo A
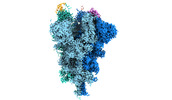
EMDB-32081: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P17
Method: single particle / : Maeda R, Fujita J, Konishi Y, Kazuma Y, Yamazaki H, Anzai I, Yamaguchi K, Kasai K, Nagata K, Yamaoka Y, Miyakawa K, Ryo A, Shirakawa K, Makino F, Matsuura Y, Inoue T, Imura A, Namba K, Takaori-Kondo A

PDB-7vq0: 
Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86
Method: single particle / : Maeda R, Fujita J, Konishi Y, Kazuma Y, Yamazaki H, Anzai I, Yamaguchi K, Kasai K, Nagata K, Yamaoka Y, Miyakawa K, Ryo A, Shirakawa K, Makino F, Matsuura Y, Inoue T, Imura A, Namba K, Takaori-Kondo A

EMDB-33666: 
chicken KNL2 in complex with the CENP-A nucleosome
Method: single particle / : Ariyoshi M, Jiang H, Makino F, Fukagawa T

EMDB-32151: 
Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1
Method: single particle / : Yoshikawa T, Makino F, Miyata T, Suzuki Y, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

PDB-7vw6: 
Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1
Method: single particle / : Yoshikawa T, Makino F, Miyata T, Suzuki Y, Tanaka H, Namba K, Sowa K, Kitazumi Y, Shirai O

EMDB-31310: 
GroEL on EG-grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T

EMDB-31311: 
GroEL on hydrophilized graphene grid (low particle density)
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T

EMDB-31312: 
A3B3 ring of V1-ATPase on EG-grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T

EMDB-31313: 
Beta-galactosidase on EG-grid
Method: single particle / : Fujita J, Makino F, Asahara H, Moriguchi M, Kumano S, Anzai I, Kishikawa J, Matsuura Y, Kato T, Namba K, Inoue T
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
